Interests
- Analysis of high-throughput biological data
- Algorithmic and analytical methods in systems/network biology
- Algorithms for data mining and analysis
- Parallel computing, algorithms for distributed systems
- Optimization problems in scientific computing
Graduate Students
- Sinan Erten, Ph.D. student
- Matthew M. Ruffalo, Ph.D. student (with T. LaFramboise)
- Marzieh Ayati, Ph.D. student
- Pavel Manaenkov, M.S. student (with P. Scacheri)
- Corey S. Adler, M.S. student
Undergraduate Students
- Ye Fang (CS)
- Theodore Roman (CS/Math, with R. Ewing)
- Alex Galante (Biology, with R. Ewing)
- Mitchell Murphy (CS, with R. Ewing)
Former Students
- Gokhan Yavas, Ph.D. (now with Case Comprehensive Cancer Center)
- Salim Akhter Chowdhury, M.S. (now with Carnegie Mellon University)
Ongoing Projects
Our research mainly focuses on development of models, algorithms,
and computational techniques to extract information from a variety of
data sources that relate to Molecular Biology, Systems Biology, and
Genetics. The main challenges associated with analyzing this type of
data include (i) the complexity of biological systems at multiple
levels (from populations to molecules), (ii) the dynamical nature of
biological phenomena in spatio-temporal dimensions, (iii) large
scale and high-dimensionality of data, along with the combinatorial
nature of interactions between different entities, and (iv) incompleteness
and noisy nature of data collected from high-throughput experiments.
While addressing these challenges, we often encounter sophisticated
abstractions and intractable computational problems, which in turn provide
us with the opportunity to contribute to computational
sciences through development of advanced algorithms and computational
techniques. The following projects are among those that are currently
undertaken by our group.
Discovery of Coordinately Dysregulated Subnetworks in Complex Phenotypes
|
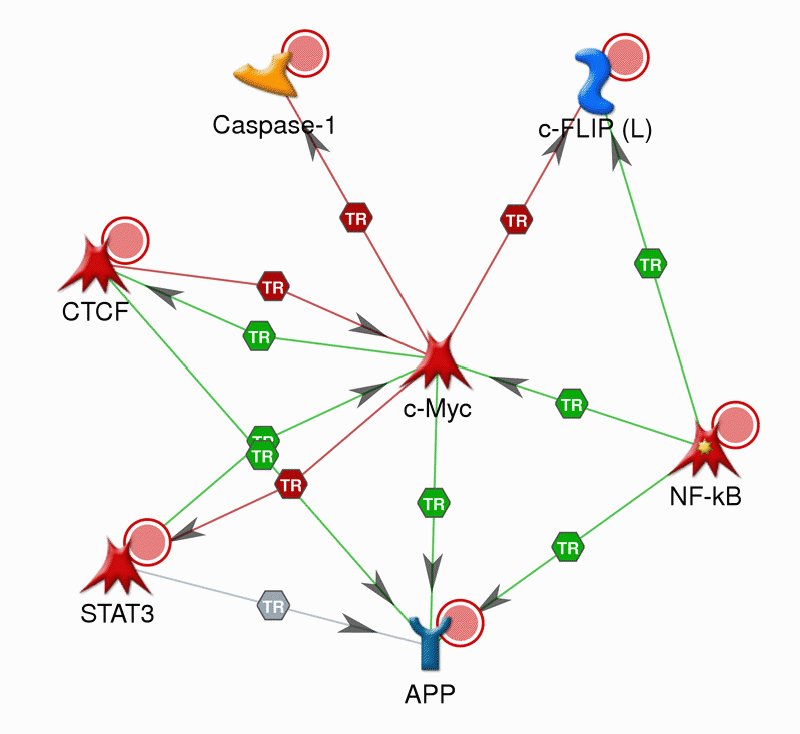 |
Cellular systems are orchestrated through combinatorial organization of thousands of biomolecules.
This complexity is reflected in the diversity of phenotypic effects, which generally present themselves as
weak signals in the expression profiles of single molecules. For this reason, researchers increasingly
focus on identification of multiple markers that together exhibit differential expression with respect
to various phenotypes. In collaboration with the research group of Mark
Chance, we focus on human colorectal cancer and develop abstractions and algorithms
to define coordinate dysregulation of multiple genes within network context and identify such network
patterns with a view to establishing markers for prognosis of cancer and targets for theurapetic
intervention. For this purpose, our algorithms integrate genomic, transcriptiomic, proteomic, and interactomic
data. This project is supported in part by NSF CAREER Award
CCF-0953195.
|
|
|
|
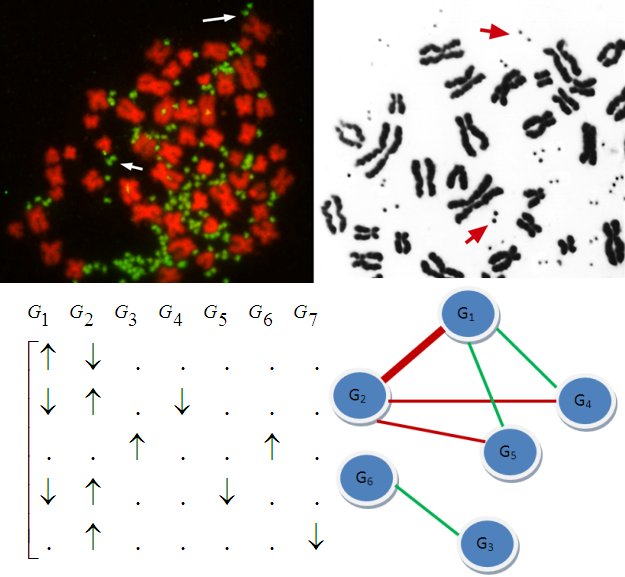
Not long ago, it was discovered that individuals may differ in copy
numbers of their genes,
meaning that a segment of DNA may have more or less copies than usual
in an individual's chromosome. Recent research suggests that
these variations are associated with many diseases including Autism
and Schizophrenia. Copy number
variation (CNV) in somatic cells also underly various cancers. Copy
numbers are usually identified using SNP microarrays, however,
short-read sequence data is emerging as an important resource
for characterizing structural variation in human genome.
In collaboration with the research group of Thomas
LaFramboise, we develop optimization based algorithms for fast and accurate identification
of rare and de novo CNVs from these two data sources, with a view
to enabling personalized genomics applications. This project is supported
by National Science Foundation
Award IIS-0916102.
|
|
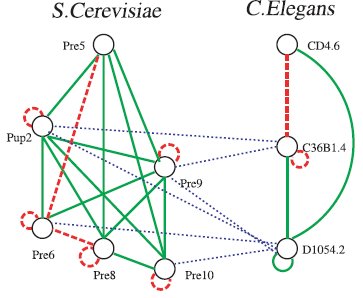
Comparative Analysis of Cellular Organization
|
|
Since all biological systems are connected to each other through
the process of evolution, a useful approach to understanding these
systems involves comparing them. The organization of cellular
systems is often abstracted using graph models that describe the
network of molecular interactions (protein-protein interactions,
gene regulation, transcription, signaling, etc.).
We develop algorithms to identify commonalities and differences
in networks that belong to different species, as well as
to reconstruct phylogenies based on network information.
|
|
Functional Annotation of Modularity in Molecular Networks
|
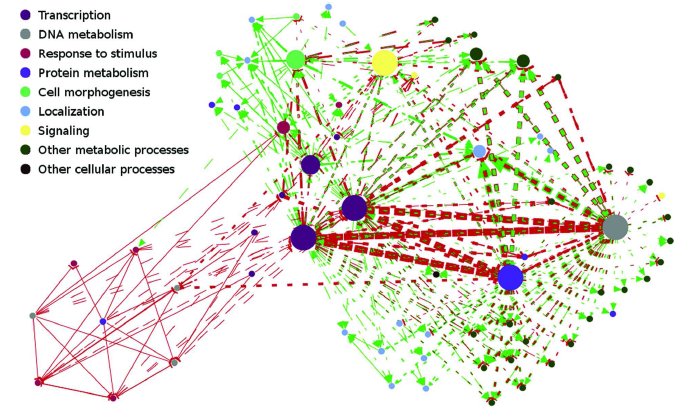 |
An important task in understanding biological systems is determining
the functions of different biological entities. Recent efforts have
been quite successful in annotating an important fraction of biomolecules,
and functional ontologies have been developed to unify our
understanding of molecular function. In collaboration with the
research group of Ananth
Grama, we develop algorithms to
extend these ontologies and annotations to systems level, by
discovering modular and overrepresented patterns in molecular
interaction networks.
|




